AI-clinician collaboration via disagreement prediction: A decision pipeline and retrospective analysis of real-world radiologist-AI interactions
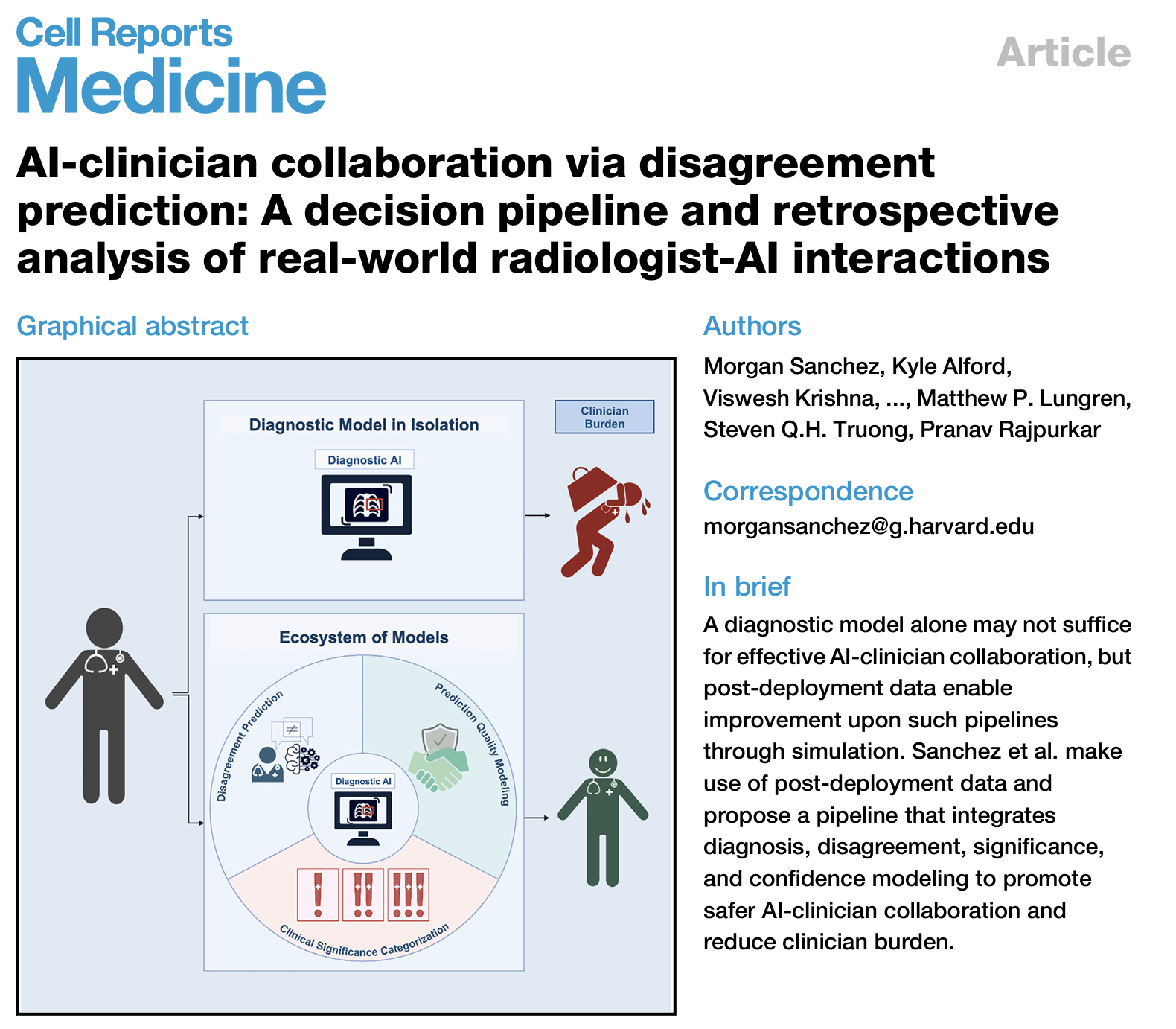
Clinical decision support tools can improve diagnostic performance or reduce variability, but they are also subject to post-deployment underperformance. Although using AI in an assistive setting offsets many concerns with autonomous AI in medicine, systems that present all predictions equivalently fail to protect against key AI safety concerns. We design a decision pipeline that supports the diagnostic model with an ecosystem of models, integrating disagreement prediction, clinical significance categorization, and prediction quality modeling to guide prediction presentation. We characterize disagreement using data from a deployed chest X-ray interpretation aid and compare clinician burden in this proposed pipeline to the diagnostic model in isolation. The average disagreement rate is 6.5%, and the expected burden reduction is 4.8%, even if 5% of disagreements on urgent findings receive a second read. We conclude that, in our production setting, we can adequately balance risk mitigation with clinician burden if disagreement false positives are reduced. Read More.


 Python (adv)
Python (adv) R (int)
R (int) MATLAB (beg)
MATLAB (beg)